EDIT-B® Clinical Validation
Making precision psychiatry a reality: a simple, rapid blood test to differentiate between depression and bipolar disorder in adults, with sensitivity and specificity in excess of 80%.
EDIT-B® Clinical Validation
Making precision psychiatry a reality: a simple, rapid blood test to differentiate between depression and bipolar disorder in adults, with sensitivity and specificity in excess of 80%.
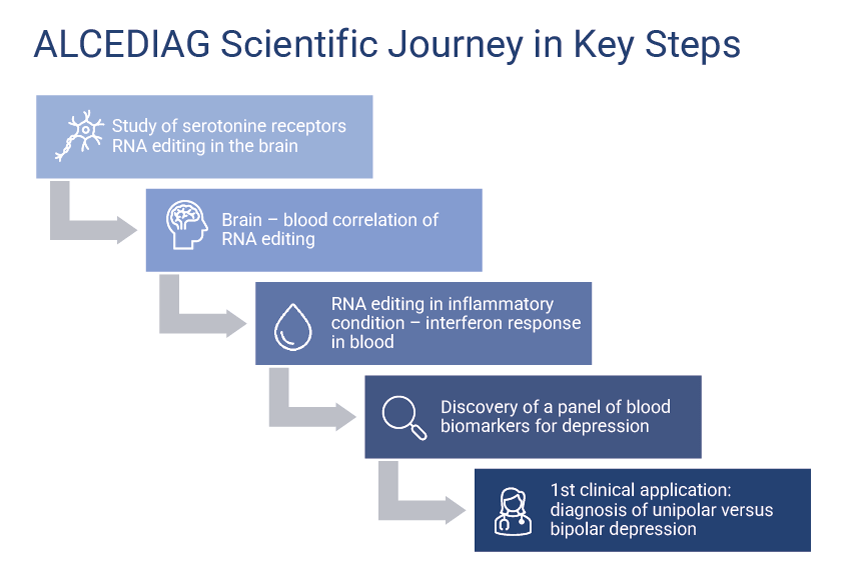
Scientific Journey
ALCEDIAG is committed to continually advancing science to better meet the needs of patients and partners. From its inception, it has embarked on a series of studies to deepen the understanding of the connections between neuropsychiatry and RNA editing, consistently exploring how these elements can be leveraged innovatively in a pre-clinical or clinical context. The key milestones of Alcediag’s scientific journey are described below, with all publications available here.
Work on Mood Disorders
Study in France – Montpellier University Hospital
Study Design
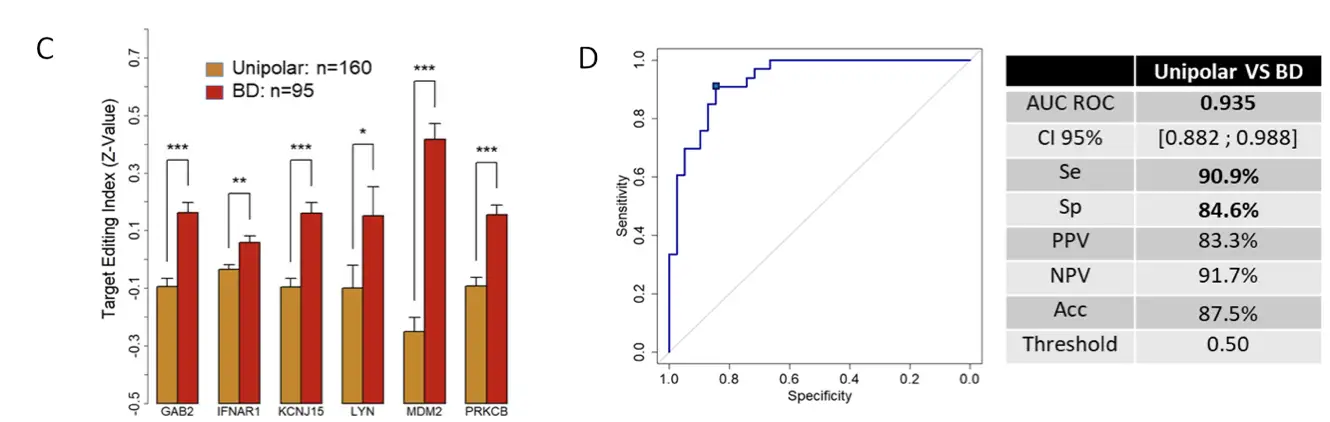
This study included 600 individuals diagnosed with depression and control subjects. The patients were recruited in Montpellier University Hospital – Department of Psychiatry. The classification of depressed patients was based on scores obtained from the Montgomery-Asberg Depression Rating Scale (MADRS) and the Inventory of Depressive Symptomatology (IDS-C30). Patients with bipolar disorder among the depressed group were evaluated by the clinician’s expertise (160 with unipolar depression, 95 with bipolar depression, and 12 with uncertain diagnosis).
Objectives
From a discovery cohort (n=57) of patients with depression and controls, 8 candidate biomarkers were selected using stringent criteria (significativity, performance, gene ontology, psychiatric disease association). The RNA editing differences for these biomarkers were compared between unipolar depressed, bipolar patients and control subjects. Using Artificial Intelligence an algorithm was created to discriminate between unipolar and bipolar depressed patients.
Limitations
Results & Key Findings
Discovery of a panel of biomarkers and algorithm: A panel of 8 RNA biomarkers involved in relevant pathways for mood disorders was identified alongside with an algorithm based on a machine learning process to differentiate unipolar depression from bipolar disorder.
Performance of the biomarkers and algorithm: The panel of eight biomarkers and algorithm were tested on a larger cohort of 255 depressed patients diagnosed with unipolar depression or bipolar disorder and showed a sensitivity of 91% and a specificity of 85%.
Validation studies and work – Montpellier University Hospital (France) & Les Toises - Psychiatry and Psychotherapy Center (Switzerland)
Study Design
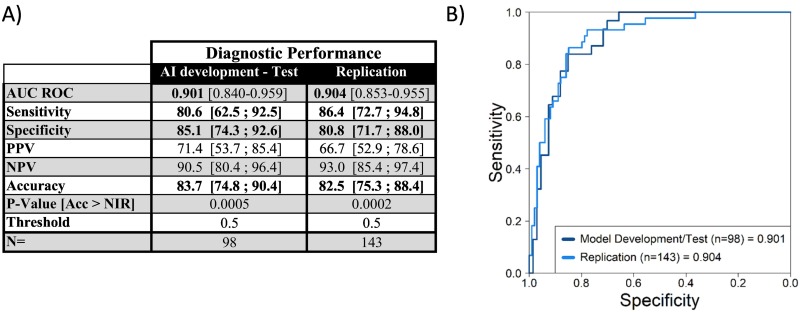
This study is a multicenter investigation involving a cohort of subjects with moderate or severe depression with a total of 143 patients enrolled (unipolar subjects, n = 100; bipolar subjects, n = 43). Classification of depressed patients was based on scores from the Montgomery-Asberg Depression Rating Scale (MADRS) and the Inventory of Depressive Symptomatology (IDS-C30), with bipolar disorder diagnosis determined by clinician expertise.
Objective
The objective was to validate the algorithm model established in an initial cohort using a second, independent cohort.
Limitations
Results & Key Findings
Biomarkers and algorithm optimization: An optimized panel of biomarkers and algorithm were developed and validated on a first cohort
Biomarkers and algorithm validation: The panel of biomarkers and algorithm were validated on a independent cohort with a performance over 80%.
Study in Brazil – Sao Paolo University (UNIFEBS) and National Institute of Translational Medicine, Ribeirao Preto, Brazil
Study Design
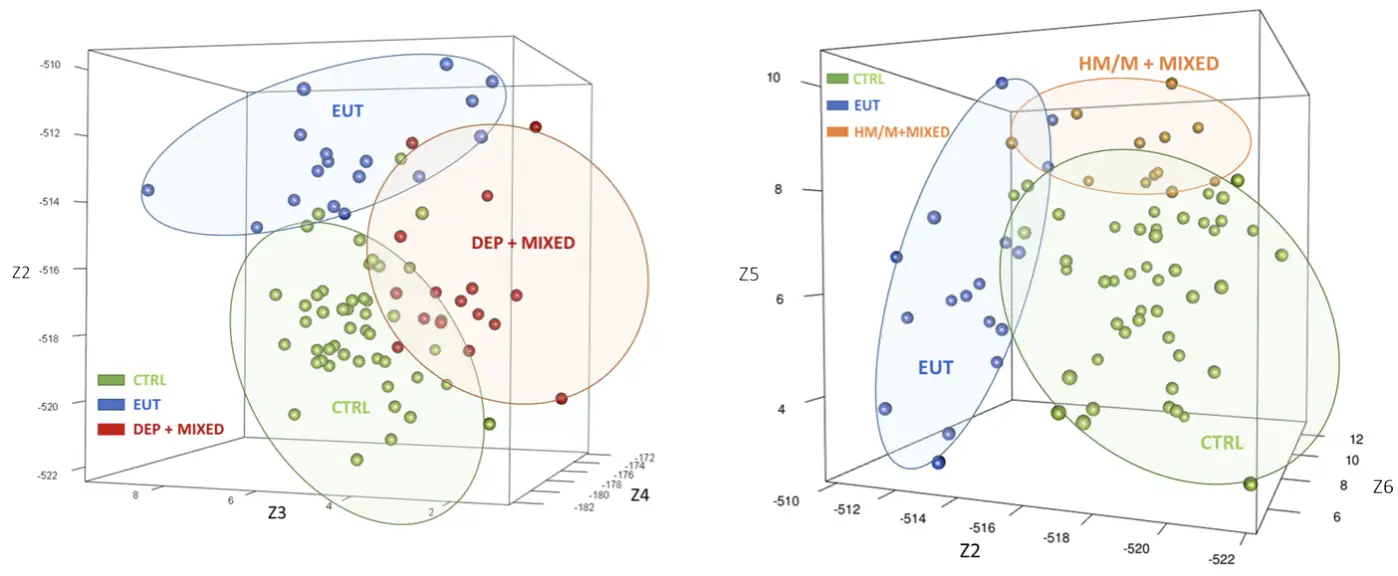
This blinded, monocenter study included 40 subjects diagnosed with bipolar disorder and 47 control subjects. Based on scores from the Young Mania Rating Scale (YMRS), Hamilton Depression Rating Scale-17 (HDRS-17), and Global Assessment of Functioning (GAF), bipolar subjects were categorized into four distinct subgroups: euthymic (n = 17), depressed (n = 11), manic/hypomanic (n = 7), and mixed (n = 5). Prior to study inclusion, bipolar subjects had maintained stable medication for a minimum of two months.
Objective
RNA editing biomarkers were compared between bipolar subgroups and control subjects. Significant biomarkers were combined, creating a unique RNA editing signature for each condition.
Limitations
This study had a limited sample size. In addition, subjects with bipolar disorder were older than controls (in mean, 45 vs. 35 years old).
Principaux résultats
RNA editing, a key mechanism for bipolar disorder: This study highlights and confirms (alongside with the study described above) RNA editing as a mechanism participating in the pathophysiology of bipolar disorder.
RNA editing and bipolar disorder in different ethnicities: This study confirms the relevance of RNA editing based biomarkers for different ethnicities than the ones performed by Alcediag in Europe.
Ongoing European multicenter Study
Study Design
This prospective multicenter study includes 436 participants diagnosed with moderate or severe depression, including individuals with unipolar depression or bipolar disorder recruited from four clinical centers in Europe: Hospital Clínic de Barcelona (Spain – Principal investigator: Dr Eduard Vieta); GHU Paris Psychiatrie & Neurosciences (France), Parc Sanitari Sant Joan de Déu (Spain), Psychiatric Center Copenhagen (Denmark). The assessment of patients with bipolar disorder within the depressed group is conducted based on the expertise of clinicians. This study is co-funded by EIT Health and the European commission. For more information, see project website.
Objective
The objective is to ensure rapid adoption of EDIT-B and obtaining the CE mark under the new EU IVDR regulation 2017/746.
Timing
The recruitment is completed.
Clinical utility study
Study Design
This study is a prospective, randomized, multicenter clinical trial conducted in France, with 450 depressed patients and a 6-month follow-up period. The primary objective is to assess the impact of the EDIT-B test on the health status of individuals with unipolar depression and bipolar disorder.
Objective
The study aims to generate valuable data for potential reimbursement of EDIT-B in France, with future considerations for other countries. This study is financed by the French Government within the Framework of France 2030 and by the European Union – Next Generation EU as part of the France relance plan.
Timing
Recruitment is ongoing.